Welcome to Virology 101- Everything you need to know!
We know that viruses harm our body. But have you ever wonder actually how do they do that? What do they look like? What disease do they cause to our body?
Well, in here, we offer the basic foundation of their classification of these viruses to the latest news to keep you up to date.
Everything you need to know, in just a click!
---
Virus-like particle vaccine could offer protection against chikungunya virus
Submitted by Mohit Joshi on Fri, 01/29/2010 - 08:36 Health News United Kingdom
London, Jan 29 : Using non-infectious virus-like particles (VLP), researchers at the National Institutes of Health have developed an experimental vaccine that could successfully protect macaques and mice against chikungunya virus.
Chikungunya virus is a mosquito-borne pathogen that has infected millions of people in Africa and Asia and causes debilitating pain.
Scientists at the National Institute of Allergy and Infectious Diseases (NIAID) developed the vaccine because there is no vaccine or treatment for chikungunya virus infection.
"Increases in global travel and trade, and possibly climate change, may be contributing to the spread of disease-carrying mosquitoes into new areas. Finding safe and effective human vaccines for chikungunya virus and other insect-borne pathogens is an important global health priority," Nature quoted NIAID Director Dr. Anthony S. Fauci as saying.
To develop the vaccine, scientists in NIAID''s Vaccine Research Center (VRC) identified the proteins that give rise to chikungunya VLPs.
The VLPs mimic actual virus particles but cannot cause infection, so they can be used safely as a vaccine to elicit immune responses.
The researchers immunized rhesus macaques with the VLPs, waited 15 weeks before exposing the animals to chikungunya virus, and observed that the vaccine provided complete protection from infection.
When the group found that antibodies were responsible for immune protection, they transferred antibody-containing serum from the vaccinated macaques to mice with deficient immune systems.
The mice then were exposed to a lethal dose of chikungunya virus, but the immune serum protected them from infection.
"This virus-like particle vaccine provides a promising way to protect against an emerging infectious disease threat. This same approach could possibly extend to viruses related to chikungunya that cause fatal diseases such as encephalitis," said VRC Director Dr. Gary Nabel.
Nabel said his group plans to seek approval for clinical trials to further evaluate the safety and efficacy of the vaccine in humans.
The details about the vaccine were published in the online version of Nature Medicine. (ANI) |
|
|
What is Virology?
Virology is the study of viruses and virus-like agents: their structure, classification and evolution,
their ways to infect and exploit cells for virus reproduction, the diseases they cause, the techniques
to isolate and culture them, and their use in research and therapy.
Virology is often considered a part of microbiology or of pathology.

Source:http://www.uib.no/imagearchive/produktbilde_lab3-600x600.jpg
Virus Classification
Virus classification involves naming and placing viruses into a taxonomic system.
It is based mainly on phenotypic characteristics, including morphology, nucleic acid
type, mode of replication, host organisms, and the type of disease they cause. A combination of two main
schemes is currently in widespread use for the classification of viruses. David Baltimore, a Nobel
Prize-winning biologist, devised the Baltimore classification system, which places viruses into one of seven
groups. These groups are designated by Roman numerals and separate viruses based on their mode of replication,
and genome type. Accompanying this broad method of classification are specific naming conventions and
further classification guidelines set out by the International Committee on Taxonomy of Viruses.
It is divided into 5 classifications: ICTV Classification, Baltimore Classification, Holmes Classification,
LHT System of Virus Classification and Subviral Agents.
1. ICTV classification
The International Committee on Taxonomy of Viruses began to devise and implement rules for the naming
and classification of viruses early in the 1990s, an effort that continues to the present day.
Viral classification starts at the level of order and follows as thus, with the taxon suffixes given in italics:
• Order (-virales)
• Family (-viridae)
• Subfamily (-virinae)
• Genus (-virus)
• Species
So far, six orders have been established by the ICTV: the Caudovirales, Herpesvirales, Mononegavirales, Nidovirales, Picornavirales, and Tymovirales.
These orders span viruses with varying host ranges:
• Caudovirales are tailed dsDNA (group I) bacteriophages,
• Herpesvirales contains large eukaryotic dsDNA viruses,
• Mononegavirales includes non-segmented (-) strand ssRNA (Group V) plant and animal viruses,
• Nidovirales is composed of (+) strand ssRNA (Group IV) viruses with vertebrate hosts,
• Picornavirales contains small (+) strand ssRNA viruses that infect a variety of plant, insect, and animal hosts
• Tymovirales contains monopartite ssRNA viruses that infect plants.
Other variations occur between the orders, for example, Nidovirales are isolated for their differentiation in expressing structural and non-structural proteins separately.
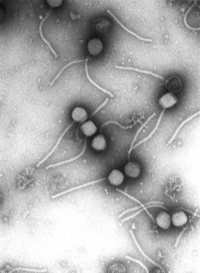
Caudovirales
Source:http://upload.wikimedia.org/wikipedia/commons/thumb/d/d0/Gamma_phage.png/200px-Gamma_phage.png
2. Baltimore classification
Baltimore classification (first defined in 1971) is a classification system that places viruses into one of seven groups depending on a combination of their nucleic acid (DNA or RNA), strandedness (single-stranded or double-stranded), Sense, and method of replication. Other classifications are determined by the disease caused by the virus
or its morphology, neither of which are satisfactory due to different viruses either causing the same disease or looking very similar. In addition, viral structures are often difficult to determine under the microscope. Classifying viruses according to their genome means that those in a given category will all behave in a similar fashion,
offering some indication of how to proceed with further research. Viruses can be placed in one of the seven following groups:
• I: dsDNA viruses (e.g. Adenoviruses, Herpesviruses, Poxviruses)
• II: ssDNA viruses (+)sense DNA (e.g. Parvoviruses)
• III: dsRNA viruses (e.g. Reoviruses)
• IV: (+)ssRNA viruses (+)sense RNA (e.g. Picornaviruses, Togaviruses)
• V: (−)ssRNA viruses (−)sense RNA (e.g. Orthomyxoviruses, Rhabdoviruses)
• VI: ssRNA-RT viruses (+)sense RNA with DNA intermediate in life-cycle (e.g. Retroviruses)
• VII: dsDNA-RT viruses (e.g. Hepadnaviruses)
2.1 DNA viruses
• Group I: viruses possess double-stranded DNA.
• Group II: viruses possess single-stranded DNA.
2.2 RNA viruses
• Group III: viruses possess double-stranded RNA genomes, e.g. rotavirus. These genomes are always segmented.
• Group IV: viruses possess positive-sense single-stranded RNA genomes. Many well known viruses are found in this group, including the picornaviruses (which is a family of viruses that includes well-known viruses like Hepatitis A virus, enteroviruses, rhinoviruses, poliovirus, and foot-and-mouth virus), SARS virus, hepatitis C virus, yellow fever virus, and rubella virus.
• Group V: viruses possess negative-sense single-stranded RNA genomes. The deadly Ebola and Marburg viruses are well known members of this group, along with influenza virus, measles, mumps and rabies.
2.3 Reverse transcribing viruses
• Group VI: viruses possess single-stranded RNA genomes and replicate using reverse transcriptase. The retroviruses are included in this group, of which HIV is a member.
• Group VII: viruses possess double-stranded DNA genomes and replicate using reverse transcriptase. The hepatitis B virus can be found in this group.
3. Holmes classification
Holmes (1948) used Carolus Linnaeus's system of binomial nomenclature to classify viruses into 3 groups under one order, Virales. They are placed as follows:
• Group I: Phaginae (attacks bacteria)
• Group II: Phytophaginae (attacks plants)
• Group III: Zoophaginae (attacks animals)
4 .LHT System of Virus Classification
The LHT System of Virus Classification is based on chemical and physical characters like nucleic acid (DNA or RNA), Symmetry (Helical or Icosahedral or Complex), presence of envelope, diameter of capsid, number of capsomers. This classification was approved by the Provisional Committee on Nomenclature of Virus (PNVC) of the International Association of Microbiological Societies (1962). It is as follows:
★ Phylum Vira (divided into 2 subphyla)>
1- Subphylum Deoxyvira (DNA viruses)
♦ Class Deoxybinala (dual symmetry)
• Order Urovirales
- Family Phagoviridae
♦ Class Deoxyhelica (Helical symmetry)
• Order Chitovirales
- Family Poxviridae
♦ Class Deoxycubica (cubical symmetry)
• Order Peplovirales
- Family Herpesviridae (162 capsomeres)
• Order Haplovirales (no envelope)
- Family Iridoviridae (812 capsomeres)
- Family Adenoviridae (252 capsomeres)
- Family Papiloviridae (72 capsomeres)
- Family Paroviridae (32 capsomeres)
- Family Microviridae (12 capsomeres)
2- Subphylum Deoxyvira (DNA viruses)
♦ Class Ribocubica
• Order Togovirales
- Family Arboviridae
• Order Lymovirales
- Family Napoviridae
- Family Reoviridae
♦ Class Ribohelica
• Order Sagovirales
- Family Stomataviridae
- Family Paramyxoviridae
- Family Myxoviridae
• Order Rbadovirales
-> Suborder Flexiviridales
- Family Mesoviridae
- Family Peptoviridae
-> Suborder Rigidovirales
- Family Pachyviridae
- Family Protoviridae
- Family Polichoviridae
5. Subviral agents
5.1 Viroids
• Family Pospiviroidae
- Genus Pospiviroid; type species: Potato spindle tuber viroid
- Genus Hostuviroid; type species: Hop stunt viroid
- Genus Cocadviroid; type species: Coconut cadang-cadang viroid
- Genus Apscaviroid; type species: Apple scar skin viroid
- Genus Coleviroid; type species: Coleus blumei viroid 1
• Family Avsunviroidae[6]
- Genus Avsunviroid; type species: Avocado sunblotch viroid
- Genus Pelamoviroid; type species: Peach latent mosaic viroid
5.2 Satellites
Satellites depend on co-infection of a host cell with a helper virus for productive multiplication. Their nucleic acids have substantially distinct nucleotide sequences from either their helper virus or host. When a satellite subviral agent encodes the coat protein in which it is encapsulated, it's then called a satellite virus.
♦ Satellite viruses[7]
• Single-stranded RNA satellite viruses
- Subgroup 1: Chronic bee-paralysis satellite virus
- Subgroup 2: Tobacco necrosis satellite virus
♦ Satellite nucleic acids
• Single-stranded satellite DNAs
• Double-stranded satellite RNAs
• Single-stranded satellite RNAs
- Subgroup 1: Large satellite RNAs
- Subgroup 2: Small linear satellite RNAs
- Subgroup 3: Circular satellite RNAs (virusoids)
5.3 Prions
Prions, named for their description as "proteinaceous and infectious particles," lack any detectable (as of 2002) nucleic acids or virus-like particles. They resist inactivation procedures that normally affect nucleic acids.[8]
• Mammalian prions:
- Agents of spongiform encephalopathies
• Fungal prions:
- PSI+ prion of Saccharomyces cerevisiae
- URE3 prion of Saccharomyces cerevisiae
- RNQ/PIN+ prion of Saccharomyces cerevisiae
- Het-s prion of Podospora anserina
The Central Dogma of Molecular Biology
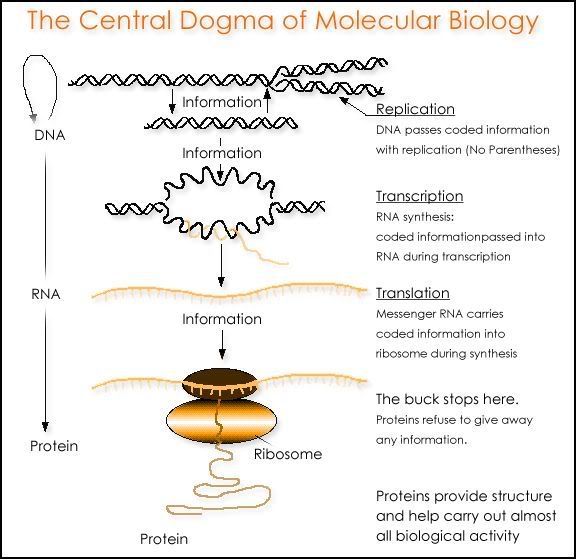
Photo from: http://www.ict-science-to-society.org/pathogenomics/images/central_dogma.gif
The central dogma of molecular biology deals with the detailed residue-by-residue transfer of sequential information. It states that information cannot be transferred back from protein to either protein or nucleic acid.
The dogma is a framework for understanding the transfer of sequence information between sequential information-carrying biopolymers, in the most common or general case, in living organisms. There are 3 major classes of such biopolymers: DNA and RNA (both nucleic acids), and protein. There are 3×3 = 9 conceivable direct transfers of information that can occur between these.
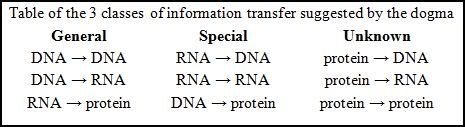
Photo captured from: http://en.wikipedia.org/wiki/Central_dogma_of_molecular_biology
The dogma classes these into 3 groups of 3: 3 general transfers (believed to occur normally in most cells), 3 special transfers (known to occur, but only under specific conditions in case of some viruses or in a laboratory), and 3 unknown transfers (believed never to occur). The general transfers describe the normal flow of biological information: DNA can be copied to DNA (DNA replication), DNA information can be copied into mRNA, (transcription), and proteins can be synthesized using the information in mRNA as a template (translation).
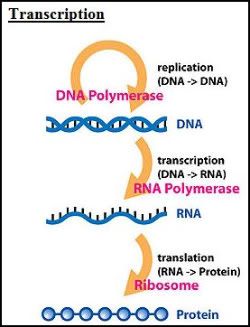
Photo: http://en.wikipedia.org/wiki/File:Central_Dogma_of_Molecular_Biochemistry_with_Enzymes.jpg
In a nutshell...
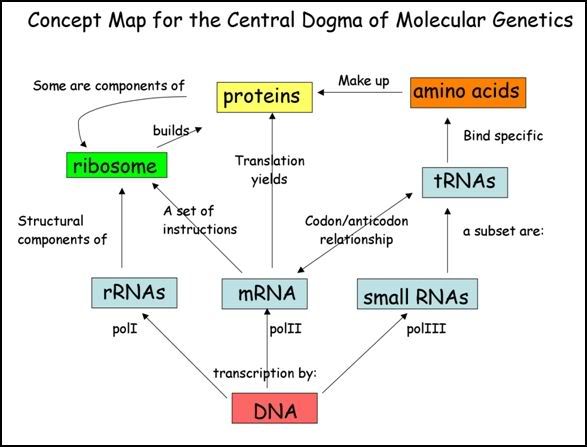
Photo from:http://www.utsc.utoronto.ca/~riggs/BGYB11/centraldogma.jpg
|

Very early form of vaccination known as variolation was developed several thousand years ago in China.
It involved the application of materials from smallpox sufferers in order to immunize others.
1717
Lady Mary Wortley Montagu observed the practice in Istanbul and tried to popularize it in Britain, but was faced with resistance.
1796
Edward Jenner developed a much safer method, using cowpox to successfully immunize a young boy against smallpox, and this practice was widely adopted.
1886
Vaccinations against other viral diseases followed, including the successful rabies vaccination by Louis Pasteur.
1890s
Dimitri Ivanovski showed that a disease of tobacco plants, tobacco mosaic disease, could be transmitted by extracts that were passed through filters fine enough to exclude even the smallest known bacteria.
In 1898, Martinus Beijerinck found that this "filterable agent" grew in the host and was thus not a mere toxin. The agent was a "living fluid" or a particle was yet to be known.
1910s
It was suggested for the first time that transduction by viruses might cause cancer.
The existence of viruses that infect bacteria (bacteriophages) was first recognized by Frederick Twort in 1911, and, independently, by Felix d'Herelle in 1917. Since bacteria could be grown easily in culture, this led to an explosion of virology research.
In 1918, French scientists showed that a "filter-passing virus" could transmit the disease to people and animals, fulfilling Koch's postulates.[2]
While plant viruses and bacteriophages can be grown comparatively easily, animal viruses normally require a living host animal, which complicates their study immensely.
1930s
it was shown that influenza virus could be grown in fertilized chicken eggs, a method that is still used today to produce vaccines.
In 1937, Max Theiler managed to grow the yellow fever virus in chicken eggs and produced a vaccine from an attenuated virus strain; this vaccine saved millions of lives and is still being used today.
Max Delbrück described the basic life cycle of a virus: instead of "growing", a virus particle is assembled from its constituent pieces in one step; eventually it leaves the host cell to infect other cells.
1960s
In 1963, the Hepatitis B virus was discovered by Baruch Blumberg who went on to develop a vaccine against Hepatitis B.
In 1965, Howard Temin described the first retrovirus: an RNA-virus that was able to insert its genome in the form of DNA into the host's genome.
A worldwide vaccination campaign led by the UN World Health Organization resulted in the eradication of smallpox in 1979.
|
|
Viruses display a wide diversity of shapes and sizes, called morphologies. Generally viruses are much smaller than bacteria. Most viruses that have been studied have a diameter between 10 and 300 nanometres. Some filoviruses have a total length of up to 1400 nm; their diameters are only about 80 nm. Most viruses cannot be seen with a light microscope so scanning and transmission electron microscopes are used to visualise virions.
To increase the contrast between viruses and the background, electron-dense "stains" are used. These are solutions of salts of heavy metals, such as tungsten, that scatter the electrons from regions covered with the stain. When virions are coated with stain (positive staining), fine detail is obscured. Negative staining overcomes this problem by staining the background only.
A complete virus particle, known as a virion, consists of nucleic acid surrounded by a protective coat of protein called a capsid. These are formed from identical protein subunits called capsomers. Viruses can have a lipid "envelope" derived from the host cell membrane. The capsid is made from proteins encoded by the viral genome and its shape serves as the basis for morphological distinction. Virally coded protein subunits will self-assemble to form a capsid, generally requiring the presence of the virus genome. Complex viruses code for proteins that assist in the construction of their capsid. Proteins associated with nucleic acid are known as nucleoproteins, and the association of viral capsid proteins with viral nucleic acid is called a nucleocapsid. The capsid and entire virus structure can be mechanically (physically) probed through atomic force microscopy
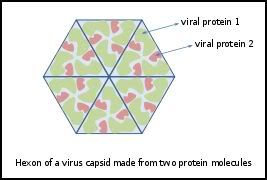
Diagram of how a virus capsid can be constructed using multiple copies of just two protein molecules
Photo from: http://en.wikipedia.org/wiki/File:Hexon.svg
• Helical
Herpes viruses have a lipid envelope
Photo from:http://upload.wikimedia.org/wikipedia/commons/1/16/Varicella_%28Chickenpox%29_Virus_PHIL_1878_lores.jpg
These viruses are composed of a single type of capsomer stacked around a central axis to form a helical structure, which may have a central cavity, or hollow tube. This arrangement results in rod-shaped or filamentous virions: these can be short and highly rigid, or long and very flexible. The genetic material, generally single-stranded RNA, but ssDNA in some cases, is bound into the protein helix by interactions between the negatively charged nucleic acid and positive charges on the protein. Overall, the length of a helical capsid is related to the length of the nucleic acid contained within it and the diameter is dependent on the size and arrangement of capsomers. The well-studied Tobacco mosaic virus is an example of a helical virus.
• Icosahedral

Electron micrograph of icosahedral adenovirus
Photo from:http://upload.wikimedia.org/wikipedia/en/f/f2/Icosahedral_Adenoviruses.jpg
Most animal viruses are icosahedral or near-spherical with icosahedral symmetry. A regular icosahedron is the optimum way of forming a closed shell from identical sub-units. The minimum number of identical capsomers required is twelve, each composed of five identical sub-units. Many viruses, such as rotavirus, have more than twelve capsomers and appear spherical but they retain this symmetry. Capsomers at the apices are surrounded by five other capsomers and are called pentons. Capsomers on the triangular faces are surround by six others and are call hexons.
• Envelope
Some species of virus envelope themselves in a modified form of one of the cell membranes, either the outer membrane surrounding an infected host cell, or internal membranes such as nuclear membrane or endoplasmic reticulum, thus gaining an outer lipid bilayer known as a viral envelope. This membrane is studded with proteins coded for by the viral genome and host genome; the lipid membrane itself and any carbohydrates present originate entirely from the host. The influenza virus and HIV use this strategy. Most enveloped viruses are dependent on the envelope for their infectivity.
• Complex
These viruses possess a capsid that is neither purely helical, nor purely icosahedral, and that may possess extra structures such as protein tails or a complex outer wall. Some bacteriophages, such as Enterobacteria phage T4 have a complex structure consisting of an icosahedral head bound to a helical tail, which may have a hexagonal base plate with protruding protein tail fibres. This tail structure acts like a molecular syringe, attaching to the bacterial host and then injecting the viral genome into the cell.
|
|

Categories of viruses by their structure:
• RNA virus - where the virus is made out of RNA
• DNA virus - where the virus is made out of DNA
• Retrovirus - a subcategory of RNA virus that makes its own DNA within the infected cell
Large category groups of viruses:
• Coronavirus
• HIV/AIDS
• Enterovirus
• Herpesvirus
• Common cold
• Flu
• Rubella
• Chicken pox
• Polio
• Rabies
• Mononucleosis
• Respiratory syncytial virus
• Dengue fever
• Rotavirus
• Viral meningitis
• West Nile fever
• Arbovirus
• Parainfluenza
• Smallpox
• Dengue hemorrhagic fever
• Viral gastroenteritis
• Acute Appendicitis
• Hepatitis A, B, C , D, E, X
• Pneumonia
• Hand, Foot, & Mouth Disease
• SARS
Coronavirus
Coronavirus is a genus of animal virus belonging to the family Coronaviridae. Coronaviruses are
enveloped viruses with a positive-sense single-stranded RNA genome and a helical symmetry. The genomic size of coronaviruses ranges from approximately 16 to 31 kilobases, extraordinarily large for an RNA virus. The name "coronavirus" is derived from the Greek κορώνα, meaning crown, as the virus envelope appears under electron microscopy (E.M.) to be crowned by a characteristic ring of small bulbous structures. This morphology is actually formed by the viral spike (S) peplomers, which are proteins that populate the surface of the virus and determine host tropism. Coronaviruses are grouped in the order Nidovirales, named for the Latin nidus, meaning nest, as all viruses in this order produce a 3' co-terminal nested set of subgenomic mRNA's during infection.

Example of coronavirus: SARS-CoV Particles
Source:http://upload.wikimedia.org/wikipedia/commons/6/6c/Sars-corona.png
HIV/AIDS
Human immunodeficiency virus (HIV) is a lentivirus (a member of the retrovirus family) that causes acquired immunodeficiency syndrome (AIDS), a condition in humans in which the immune system begins to fail, leading to life-threatening opportunistic infections. Infection with HIV occurs by the transfer of blood, semen, vaginal fluid, pre-ejaculate, or breast milk. Within these bodily fluids, HIV is present as both free virus particles and virus within infected immune cells. The four major routes of transmission are unsafe sex, contaminated needles, breast milk, and transmission from an infected mother to her baby at birth (Vertical transmission).
Screening of blood products for HIV has largely eliminated transmission through blood transfusions or infected blood products in the developed world.
HIV infects primarily vital cells in the human immune system such as helper T cells (to be specific, CD4+ T cells), macrophages, and dendritic cells. HIV infection leads to low levels of CD4+ T cells through three main mechanisms: First, direct viral killing of infected cells; second, increased rates of apoptosis in infected cells; and third, killing of infected CD4+ T cells by CD8 cytotoxic lymphocytes that recognize infected cells. When CD4+ T cell numbers decline below a critical level, cell-mediated immunity is lost, and the body becomes progressively more susceptible to opportunistic infections.
Most people infected with HIV eventually develop AIDS. These individuals mostly die from opportunistic infections or malignancies associated with the progressive failure of the immune system. HIV progresses to AIDS at a variable rate affected by viral, host, and environmental factors; HIV-specific treatment delays this process. Most will progress to AIDS within 10 years of HIV infection: some will have progressed much sooner, and some will take much longer. Treatment with anti-retrovirals increases the life expectancy of people infected with HIV. Even after HIV has progressed to diagnosable AIDS, the average survival time with antiretroviral therapy was estimated to be more than 5 years as of 2005. Without antiretroviral therapy, someone who has AIDS typically dies within a year.
HIV Virus infecting a cell
Photo by: http://www.aps.org/publications/apsnews/200705/images/aids_photo_web.jpg
Enterovirus
The enteroviruses are a genus of (+)ssRNA viruses associated with several human and mammalian diseases. Historically the most significant has been the Poliovirus. Other types are coxsackie and echovirus.
Enterovirus are the most common cause of aseptic meningitis and can cause serious disease especially in infants and the immunocompromised.
Human enteroviruses (family Picornaviridae) infect millions of people worldwide each year, resulting in a wide range of clinical outcomes ranging from unapparent infection to mild respiratory illness (common cold), hand, foot and mouth disease, acute hemorrhagic conjunctivitis, aseptic meningitis, myocarditis, severe neonatal sepsis-like disease, and acute flaccid paralysis. On the basis of their pathogenesis in humans and experimental animals, the enteroviruses were originally classified into four groups, polioviruses, coxsackie A viruses (CA), coxsackie B viruses (CB), and echoviruses, but it was quickly realized that there were significant overlaps in the biological properties of viruses in the different groups. The more recently isolated enteroviruses have been named with a system of consecutive numbers: EV68, EV69, EV70, and EV71.
There are 62 non-polio enteroviruses that can cause disease in humans: 23 Coxsackie A viruses, 6 Coxsackie B viruses, 28 echoviruses, and 5 other enteroviruses.

Enterovirus usually reproduce in the intestine.
Photo from: http://www.worsleyschool.net/science/files/virus/enterovirus.jpg
Enterovirus also cause petechial rash in babies.

Photo from: http://www.baby-medical-questions-and-answers.com/images/meningococcaemia3.jpg
Flu

Photo from: http://virologyness.files.wordpress.com/2009/02/spl_5_m130797-man_sneezing.jpg
Influenza, commonly referred to as the flu, is an infectious disease caused by RNA viruses of the family Orthomyxoviridae (the influenza viruses), that affects birds and mammals. The most common symptoms of the disease are chills, fever, sore throat, muscle pains, severe headache, coughing, weakness/fatigue and general discomfort. Sore throat, fever and coughs are the most frequent symptoms. In more serious cases, influenza causes pneumonia, which can be fatal, particularly for the young and the elderly. Although it is often confused with other influenza-like illnesses, especially the common cold, influenza is a much more severe disease than the common cold and is caused by a different type of virus. Influenza may produce nausea and vomiting, particularly in children, but these symptoms are more common in the unrelated gastroenteritis, which is sometimes called "stomach flu" or "24-hour flu".
Typically, influenza is transmitted through the air by coughs or sneezes, creating aerosols containing the virus. Influenza can also be transmitted by direct contact with bird droppings or nasal secretions, or through contact with contaminated surfaces. Airborne aerosols have been thought to cause most infections, although which means of transmission is most important is not absolutely clear. Influenza viruses can be inactivated by sunlight, disinfectants and detergents.
As the virus can be inactivated by soap, frequent hand washing reduces the risk of infection. The most common human vaccine is the trivalent influenza vaccine (TIV) that contains purified and inactivated material from three viral strains. Typically, this vaccine includes material from two influenza A virus subtypes and one influenza B virus strain.
Rubella
Rubella, commonly known as German measles, is a disease caused by the rubella virus. The name "rubella" is derived from the Latin, meaning little red. Rubella is also known as German measles because the disease was first described by German physicians in the mid-eighteenth century. This disease is often mild and attacks often pass unnoticed. The disease can last one to three days. Children recover more quickly than adults. Infection of the mother by Rubella virus during pregnancy can be serious; if the mother is infected within the first 20 weeks of pregnancy, the child may be born with congenital rubella syndrome (CRS), which entails a range of serious incurable illnesses. Spontaneous abortion occurs in up to 20% of cases
Chicken pox
Chickenpox is a highly contagious illness caused by primary infection with varicella zoster virus (VZV). It usually starts with vesicular skin rash mainly on the body and head rather than at the periphery and become itchy raw pockmarks which mostly heal without scarring.
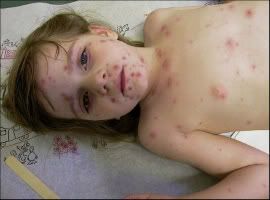 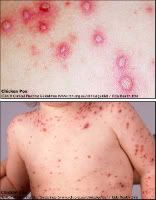
Photo(left) from: http://123shadow.files.wordpress.com/2009/03/chicken-pox.jpg
Photo(right) from: http://atheistage.org/wp-content/uploads/2009/01/chicken-pox.jpg
Chicken pox is spread easily through coughs or sneezes of ill individuals, or through direct contact with secretions from the rash.
Chickenpox is rarely fatal, although it is generally more severe in adult males than in adult females or children. Pregnant women and those with a suppressed immune system are at highest risk of serious complications. The most common late complication of chicken pox is shingles, caused by reactivation of the varicella zoster virus decades after the initial episode of chickenpox. Chickenpox has been observed in other primates.
Rabies
Rabies is a viral neuroinvasive disease that causes acute encephalitis (inflammation of the brain) in warm-blooded animals. It is zoonotic (i.e., transmitted by animals), most commonly by a bite from an infected animal but occasionally by other forms of contact. Rabies is almost invariably fatal if post-exposure prophylaxis is not administered prior to the onset of severe symptoms. It is a significant killer of livestock in some countries.
The rabies virus travels to the brain by following the peripheral nerves. The incubation period of the disease depends on how far the virus must travel to reach the central nervous system, usually taking a few months. Once the infection reaches the central nervous system and symptoms begin to show, the infection is practically untreatable and usually fatal within days.
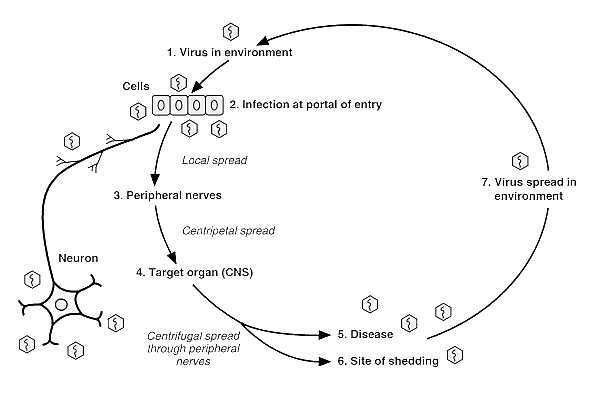
Entry of rabies virus into body
Photo from: http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=mmed&part=A2413
Hand, Foot, & Mouth Disease
Hand, foot and mouth disease (HFMD) is a human syndrome caused by intestinal viruses of the Picornaviridae family. The most common strains causing HFMD are Coxsackie A virus and Enterovirus 71 (EV71). HFMD usually affects infants and children, and is quite common. It is moderately contagious and is spread through direct contact with the mucus, saliva, or feces of an infected person. It typically occurs in small epidemics in nursery schools or kindergartens, usually during the summer and autumn months. The usual incubation period is 3–7 days.
 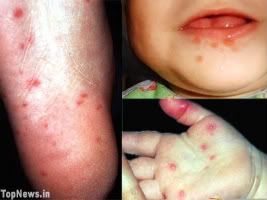
(Left)Coxsackie A virus
(Right) Hand, Foot & Mouth Disease
Photo(left) from: http://stevejensen.smugmug.com/photos/500917764_LVKsa-M.jpg
Photo(right) from: http://www.topnews.in/health/files/HFMD.jpg
Viral gastroenteritis
Gastroenteritis (also known as gastro, gastric flu, tummy bug in some countries, and stomach flu, although unrelated to influenza) is inflammation of the gastrointestinal tract, involving both the stomach and the small intestine (see also gastritis and enteritis) and resulting in acute diarrhea. The inflammation is caused most often by an infection from certain viruses or less often by bacteria, their toxins, parasites, or an adverse reaction to something in the diet or medication. Worldwide, inadequate treatment of gastroenteritis kills 5 to 8 million people per year, and is a leading cause of death among infants and children under 5.
At least 50% of cases of gastroenteritis due to foodborne illness are caused by norovirus. Another 20% of cases, and the majority of severe cases in children, are due to rotavirus. Other significant viral agents include adenovirus and astrovirus.
Different species of bacteria can cause gastroenteritis, including Salmonella, Shigella, Staphylococcus, Campylobacter jejuni, Clostridium, Escherichia coli, Yersinia, and others. Some sources of the infection are improperly prepared food, reheated meat dishes, seafood, dairy, and bakery products. Each organism causes slightly different symptoms but all result in diarrhea. Colitis, inflammation of the large intestine, may also be present.

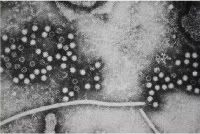
Electron Micrographs of viruses that cause gastroenteritis in humans.
A = rotavirus, B = adenovirus, C = norovirus and D = astrovirus. Magnification: 200,000x.
Photo from: http://upload.wikimedia.org/wikipedia/en/7/71/Gastroenteritis_viruses.jpg
Severe Acute Respiratory Syndrome (SARS)
Severe Acute Respiratory Syndrome is a respiratory disease in humans which is caused by the SARS coronavirus (SARS-CoV).
Initial electron microscopic examination in Hong Kong and Germany found viral particles with structures suggesting paramyxovirus in respiratory secretions of SARS patients. The Pasteur Institute in Paris identified coronavirus in samples taken from six patients, so did the laboratory of Malik Peiris at the University of Hong Kong, which in fact was the first to announce (on 21 March 2003) the discovery of a new coronavirus as the possible cause of SARS after successfully cultivating it from tissue samples and was also amongst the first to develop a test for the presence of the virus.
Coronavirus (CoV) genome replication takes place in the cytoplasm in a membrane-protected microenvironment and starts with the translation of the genome to produce the viral replicase. CoV transcription involves a discontinuous RNA synthesis (template switch) during the extension of a negative copy of the subgenomic mRNAs. The requirement for base pairing during transcription has been formally demonstrated in arteriviruses and CoVs. The CoV N protein is required for coronavirus RNA synthesis and has RNA chaperon activity that may be involved in template switch. Both viral and cellular proteins are required for replication and transcription. CoVs initiate translation by cap-dependent and cap-independent mechanisms. Cell macromolecular synthesis may be controlled after CoV infection by locating some virus proteins in the host cell nucleus.
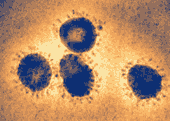
SARS coronavirus (SARS-CoV), the causative agent of the syndrome
Photo from: http://upload.wikimedia.org/wikipedia/commons/6/6c/Sars-corona.png
Infection by different coronaviruses cause in the host alteration in the transcription and translation patterns, in the cell cycle, the cytoskeleton, apoptosis and coagulation pathways, inflammation and immune and stress responses. The balance between genes up- and down-regulated could explain the pathogenesis caused by these viruses. Coronavirus expression systems based on single genome constructed by targeted recombination, or by using infectious cDNAs, have been developed. The possibility of expressing different genes under the control of transcription regulating sequences (TRSs) with programmable strength and engineering tissue and species tropism indicates that CoV vectors are flexible. CoV based vectors have emerged with high potential vaccine development and possibly for gene therapy.
Hepatitis A, B, C , D, E, X
-A
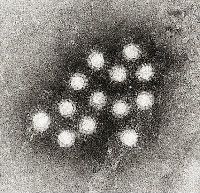
Electron micrograph of the Hepatitis A virus (HAV)
Photo from:http://upload.wikimedia.org/wikipedia/commons/f/f5/Hepatitis_A_virus_02.jpg
Hepatitis A (formerly known as infectious hepatitis) is an acute infectious disease of the liver caused by the hepatitis A virus (HAV), which is most commonly transmitted by the fecal-oral route via contaminated food or drinking water. Every year, approximately 10 million people worldwide are infected with the virus. The time between infection and the appearance of the symptoms, (the incubation period), is between two and six weeks and the average incubation period is 28 days.
-B
Hepatitis B is a disease caused by hepatitis B virus (HBV) which infects the liver of hominoidae, including humans, and causes an inflammation called hepatitis. Originally known as "serum hepatitis",the disease has caused epidemics in parts of Asia and Africa, and it is endemic in China. About a third of the world's population, more than 2 billion people, have been infected with the hepatitis B virus. This includes 350 million chronic carriers of the virus. Transmission of hepatitis B virus results from exposure to infectious blood or body fluids containing blood.
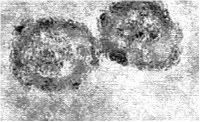
Electron micrograph of Hepatitis B virus particles and surface antigen from blood.
Photo from:http://upload.wikimedia.org/wikipedia/commons/3/36/Hepatitis_B_virus_1.jpg
Hepatitis B virus is an hepadnavirus—hepa from hepatotrophic and dna because it is a DNA virus—and it has a circular genome composed of partially double-stranded DNA. The viruses replicate through an RNA intermediate form by reverse transcription, and in this respect they are similar to retroviruses. Although replication takes place in the liver, the virus spreads to the blood where virus-specific proteins and their corresponding antibodies are found in infected people. Blood tests for these proteins and antibodies are used to diagnose the infection.
-C
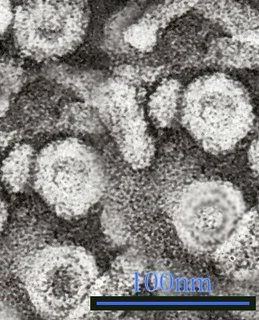
Electron micrograph of Hepatitis C virus
Photo from:http://upload.wikimedia.org/wikipedia/commons/d/dc/Em_flavavirus-HCV_samp1c.jpg
Hepatitis C is an infectious disease affecting the liver, caused by the hepatitis C virus (HCV). The infection is often asymptomatic, but once established, chronic infection can progress to scarring of the liver (fibrosis), and advanced scarring (cirrhosis) which is generally apparent after many years. In some cases, those with cirrhosis will go on to develop liver failure or other complications of cirrhosis, including liver cancer. Hepatitis C virus (HCV) is a small (55-65 nm in size), enveloped, positive sense single strand RNA virus in the family Flaviviridae. Although Hepatitis A virus, Hepatitis B virus, and Hepatitis C virus have similar names (because they all cause liver inflammation), these are distinctly different viruses both genetically and clinically.
-D
Hepatitis D, also referred to as Hepatitis D virus (HDV) and classified as Hepatitis delta virus, is a disease caused by a small circular RNA virus. HDV is considered to be a subviral satellite because it can propagate only in the presence of the Hepatitis B virus (HBV). Transmission of HDV can occur either via simultaneous infection with HBV (coinfection) or via infection of an individual previously infected with HBV (superinfection).
-E
Hepatitis E is a viral hepatitis (liver inflammation) caused by infection with a virus called hepatitis E virus (HEV). HEV is a positive-sense single-stranded RNA icosahedral virus with a 7.5 kilobase genome. Although it was originally classified in the Caliciviridae family, the virus has since been classified into the genus Hepevirus, but was not assigned to a viral family. The virus itself is a small non-enveloped particle.
The genome is approximately 7200 bases in length, is a polyadenylated single-strand RNA molecule that contains three discontinuous and partially overlapping open reading frames (ORFs) along with 5' and 3' cis-acting elements, which have important roles in HEV replication and transcription. ORF1 encode a methyltransferase, protease, helicase and replicase; ORF2 encode the capsid protein and ORF3 encodes a protein of undefined function. A three-dimensional, atomic-resolution structure of the capsid protein in the context of a virus-like particle has been described.
Hepatitis E virus (HEV), the major etiologic agent of enterically transmitted non-A, non-B hepatitis worldwide
Photo from:http://patric.vbi.vt.edu/organism/overview/images/7.jpg
-X
Hepatitis X, also known as the "Lost Hepatitis" is an acute infectious disease of the liver caused by Hepatitis virus, which is most commonly transmitted by the fecal-oral route via contaminated food or drinking waterOften the virus will show itself, but later the patient will lose all symptoms. Roughly one week later the virus will show itself again stronger and more dangerously. Early symptoms of hepatitis X infection can be mistaken for influenza, but some sufferers, especially children, exhibit no symptoms at all. Symptoms typically appear 1 to 2 weeks, (the incubation period ), after the initial infection.
|

Viral replication is the term used by virologists to describe the formation of biological viruses during the infection process in the target host cells. Viruses must first get into the cell before viral replication can occur. From the perspective of the virus, the purpose of viral replication is to allow production and survival of its kind. By generating abundant copies of its genome and packaging these copies into viruses, the virus is able to continue infecting new hosts.
Lytic cycle
The lytic cycle is one of the two cycles of viral reproduction, the other being the lysogenic cycle. The lytic cycle is typically considered the main method of viral replication, since it results in the destruction of the infected cell.
Viruses of the lytic cycle are called virulent viruses. The lytic cycle is a six-stage cycle. In the first stage, called "penetration," the virus injects its own nucleic acids into a host cell. Then the viral acids form a circle in the center of the cell. The cell then mistakenly copies the viral acids instead of its own nucleic acids. Then the viral DNA organize themselves as viruses inside the cell. When the number of viruses inside becomes too much for the cell to hold, the membrane splits and the viruses are free to infect other cells.
Lysogenic Cycle
Lysogeny, or the lysogenic cycle, is one of two phases of viral reproduction (the lytic cycle is the other). Lysogeny is characterized by integration of the bacteriophage nucleic acid into the host bacterium's genome. The newly integrated genetic material, called a prophage can be transmitted to daughter cells at each subsequent cell division, and a later event (such as UV radiation) can release it, causing proliferation of new phages via the lytic cycle. Lysogenic cycles can also occur in eukaryotes, although the method of incorporation of DNA is not fully understood.
Lysogenic conversion
In some interactions between lysogenic phages and bacteria, lysogenic conversion may occur. It is when a temperate phage induces a change in the phenotype of the bacteria infected that is not part of a usual phage cycle. Changes can often involve the external membrane of the cell by making it impervious to other phages or even by increasing the pathogenic capability of the bacteria for a host.
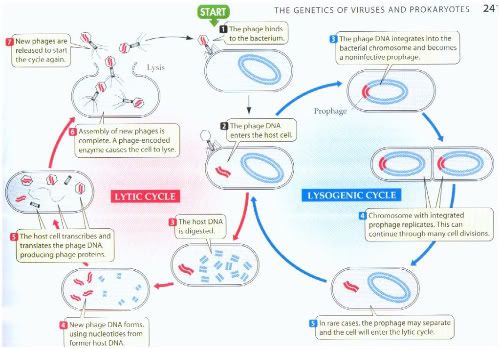
Photo from:http://www.prism.gatech.edu/~gh19/b1510/8lytic.jpg
---
Virus- Host Interactions
Initiation Phase:
Attachment, Penetration and Uncoating
The virion protein or antireceptor binds to a cell surface receptor. A classic example of this process is the haemaglutinin antireceptor of influenza virus. Complex viruses such as pox or herpes may have more than one antireceptor. Furthermore, the antireceptor may have several domains which react with different receptors.
Cellular receptors are largely glycoproteins. The interaction of the receptor and antireceptor requires counter ions to reduce electrostatic repulsions and is temperature and energy independent. In some cases, attachment leads to irreversible changes in the structure of the virion. The influenza haemaglutinin protein bind to glycoproteins carrying neuraminic (sialic) acid. The virus also has a neuraminidase on its surface and can detach from the cell by hydrolysing neuraminic acid from the glycoprotein.
Entry of unenveloped viruses
The uptake of unenveloped viruses was, until recently, always considered to involve endocytosis into an endosome, i.e. to be pH-dependent and sensitive to ionophores and lysosomotropic agents. An example is the poliovirus which belongs to the picornavirus family. This is an icosahedral T=3 virus.
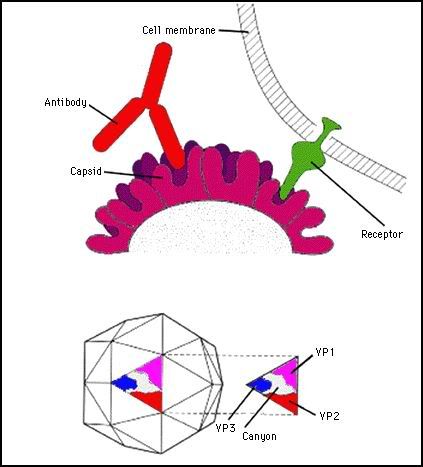
Photo from:http://www.microbiologybytes.com/virology/3035pics/3035Rep4.gif
The capsid consists of 4 polypeptides, VP1, 2, 3 and 4.
VP4 is buried in the capsid and associated with the viral RNA.
VP1, 2 and 3 form the external capsid. Five VP1 protein subunits at the 5 fold axis of symmetry form a canyon in the capsid which is the recognition site for the receptor.
The polio receptor belongs to IgG superfamily (as does CD4), it has a MW of 45Kd. It has an amino terminal Ig-like domain which binds to the virus, a transmembrane domain and a cytoplasmic domain (which is dispensible for function.There are several thousand copies of the receptor present on a cell.
After binding to the receptor, the virus is endocytosed into an endosome. At a low pH, poliovirus becomes more lipophilic and can associate with the endosome membrane possibly forming a pore. Presumably the pH shift results in exposure of hydrophobic domains in the capsid proteins. During this process VP4 is lost from the particle; VP2 may also be lost. This gives the particle increased flexibility. The genomic RNA is ejected through an endosomal pore into the cytoplasm.
Uncoating
This is the general term applied to events after penetration which allow the virus to express its genome. It is poorly understood.
In reoviruses, the capsid only ever partially disintegrates and replication takes place in a structured particle.
In poxviruses, host factors induce the disruption of the virus. The release of DNA from the core depends upon viral factors made after entry.
Orthomyxoviruses, paramyxoviruses and picornaviruses all lose the protective envelope or capsid upon entry into the cytoplasm. In the influenza virus an envelope viral protein called M2 may allow endosomal protons into the virion particle resulting in its partial dissolution and permitting replication. M2 is a polypeptide of 97aa It forms a tetrameric channel forming structure within the viral envelope. Amantadine and rimantidine are anti-influenza drugs which function in part by inhibiting M2. In cells treated with rimantidine the nucleocapsid remains associated with matrix protein and does not go to the nucleus.
In herpesviruses, adenoviruses and polyomaviruses, the capsid is eventually routed along the cytoskeleton to nuclear envelope.
Replication
In retroviruses, the matrix protein probably stays associated with cytoplasmic membrane after entry. Reverse transcription ie copying the RNA genome into a double stranded DNA form is thought to take place in a structured remnant of the capsid in the cytoplasm. The enzyme concerned is reverse transcriptase. It is both an RNA and DNA directed DNA polymerase. It also has an associated RNAse activity. The process is illustrated below:
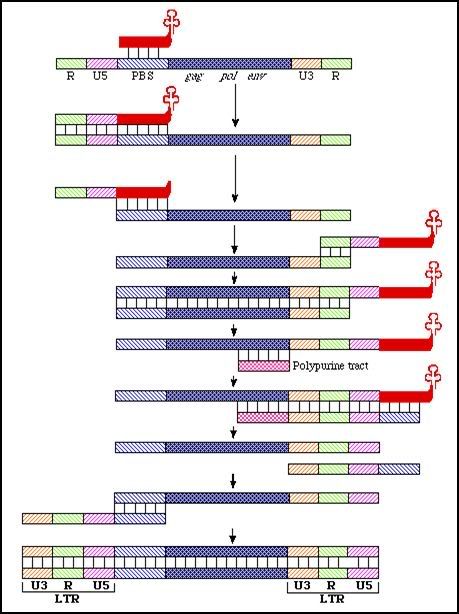
Photo from:http://www.microbiologybytes.com/virology/3035pics/Retro5.gif
It results in the formation of a double-stranded proviral DNA which is longer than the viral RNA by one copy of U3, R and U5. In reality, the procedure is more complicated than shown. There are probably more than 2 strand jumps because retroviral RNA is often broken or nicked. At every nick of necessity the RT complex has to switch to another RNA strand (remember that there are two present).
The proviral DNA is transported to the nucleus and integrated into the cellular DNA. HIV is not thought to show any obvious preferences in the sites of integration. The viral enzyme catalyzing this process is called integrase.
(Info link: http://www.microbiologybytes.com/virology/3035Replication.html)
Assembly, maturation and release
Some viruses make morphogenetic factors which are not structurally part of the virus but whose presence is required for normal assembly. These are sometimes called molecular chaperones. Cellular chaperones may also take part in this process.
Other viruses self-assemble. It has been known for more than 40 years that a mixture of TMV proteins and RNA will assemble and make infectious virus in the test tube. This suggests that the TMV structure is a minimum free energy state for its constituent parts.
Symmetry is important in viral assembly. It reduces ambiguities in the assembly process and makes it easier. If the coat protein had to form different contacts in different places within the virus structure it would lead to ambiguities in assembly.
In the assembly of TMV the coat protein first forms a disc structure with 17 subunits per ring. This is close to the 16.34 subunits per turn found in the virus. Close examination of electron micrographs shows that the discs are not actually symmetrical, they have a pronounced polarity:
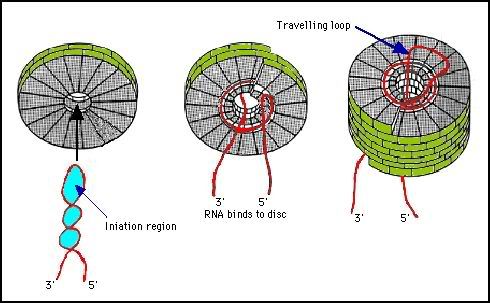
Photo from:http://www.microbiologybytes.com/virology/3035pics/3035Rep12.gif
Budding
Some viruses, e.g. herpesviruses, assemble in the nucleus. They acquire an envelope as they pass through the inner nuclear membrane.
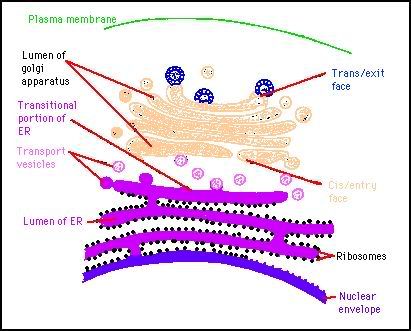
Photo from:http://www.microbiologybytes.com/virology/3035pics/3035Rep14.gif
They then accumulate, between the inner and outer lamella of the nucleus. This is topologically equivalent to the cisternae of the cytoplasmic reticulum. From here they pass in a vesicle to the cell surface. These viruses are protected from the cytoplasm. This budding process is invariably cytolytic.
Other enveloped viruses acquire their lipid bilayer from the plasma membrane. Capsid assembly can take place in the cytoplasm or in the case of HIV at the plasma membrane. The HIV gag fusion protein p55 is inserted into the lipid membrane by because of a myristyl group at the N-terminus. A patch of gag proteins form associated with the membrane either because of protein-protein interactions or because of association with the viral RNA. The approximately spherical virus may form because of a curvature induced by the gag protein association or because of the helical nature of the RNA. The RNA becomes extensively associated with the nucleocapsid moiety of gag.
(Info link: http://www.microbiologybytes.com/virology/3035Replication.html)
Virus- Host Interaction in a nutshell...
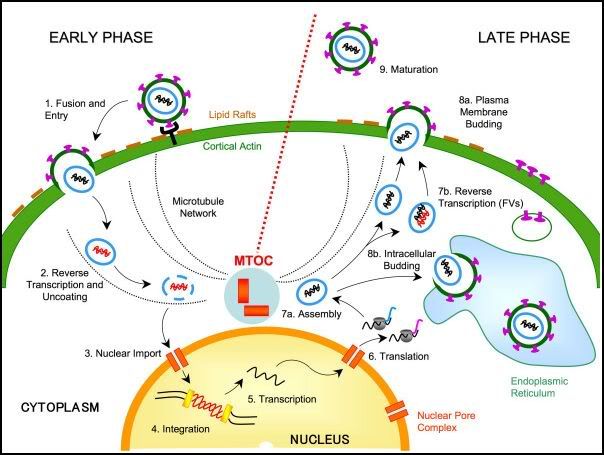
Photo from:http://project0802b.files.wordpress.com/2009/02/1742-4690-4-27-2.jpg
|
|
1. Virus Isolation
Viruses are obligate intracellular parasites that require living cells in order to replicate. Cultured cells, eggs and laboratory animals may be used for virus isolation. Although embroyonated eggs and laboratory animals are very useful for the isolation of certain viruses, cell cultures are the sole system for virus isolation in most laboratories. The development of methods for cultivating animal cells has been essential to the progress of animal virology.
To prepare cell cultures, tissue fragments are first dissociated, usually with the aid of trypsin or collagenase. The cell suspension is then placed in a flat-bottomed glass or plastic container (petri dish, a flask, a bottle, test tube) together with a suitable liquid medium. e.g. Eagle's, and an animal serum. After a variable lag, the cells will attach and spread on the bottom of the container and then start dividing, giving rise to a primary culture. Attachment to a solid support is essential for the growth of normal cells.
Primary and Secondary Cultures
Primary cultures are maintained by changing the fluid 2 or 3 times a week. When the cultures become too crowded, the cells are detached from the vessel wall by either trypsin or EDTA, and portions are used to initiate secondary cultures. In both primary and secondary cultures, the cells retain some of the characteristics of the tissue from which they are derived.
Maintaining cells in culture
Cells are grown and maintained at an appropriate temperature and gas mixture (typically, 37°C, 5% CO2 for mammalian cells) in a cell incubator. Culture conditions vary widely for each cell type, and variation of conditions for a particular cell type can result in different phenotypes being expressed.
Aside from temperature and gas mixture, the most commonly varied factor in culture systems is the growth medium. Recipes for growth media can vary in pH, glucose concentration,growth factors, and the presence of other nutrients. The growth factors used to supplement media are often derived from animal blood, such as calf serum. One complication of these blood-derived ingredients is the potential for contamination of the culture with viruses or prions, particularly in biotechnology medical applications. Current practice is to minimize or eliminate the use of these ingredients wherever possible, but this cannot always be accomplished.
Cells can be grown in suspension or adherent cultures. Some cells naturally live in suspension, without being attached to a surface, such as cells that exist in the bloodstream. There are also cell lines that have been modified to be able to survive in suspension cultures so that they can be grown to a higher density than adherent conditions would allow.
Cell Strains and Cell Lines
Cells from primary cultures can often be transferred serially a number of times. The cells may then continue to multiply at a constant rate over many successive transfers. Eventually, after a number of transfers, the cells undergo culture senescence and cannot be transferred any longer. For human diploid cell cultures, the growth rate declines after about 50 duplications. During the multiplication of the cell strain, some cells become altered in that they acquire a different morphology, grow faster, and become able to start a cell culture from a smaller number of cells. These cells are immortalized and have an unlimited life-span. However, they retain contact inhibition.
Cell line cross-contamination
Cell line cross-contamination can be a problem for scientists working with cultured cells. Studies suggest that anywhere from 15–20% of the time, cells used in experiments have been misidentified or contaminated with another cell line. Problems with cell line cross contamination have even been detected in lines from the NCI-60 panel, which are used routinely for drug-screening studies. Major cell line repositories including the American Type Culture Collection (ATCC) and the German Collection of Microorganisms and Cell Cultures (DSMZ) have received cell line submissions from researchers that were misidentified by the researcher. Such contamination poses a problem for the quality of research produced using cell culture lines, and the major repositories are now authenticating all cell line submissions. ATCC uses short tandem repeat (STR) DNA fingerprinting to authenticate its cell lines.
To address this problem of cell line cross-contamination, researchers are encouraged to authenticate their cell lines at an early passage to establish the identity of the cell line. Authentication should be repeated before freezing cell line stocks, every two months during active culturing and before any publication of research data generated using the cell lines. There are many methods for identifying cell lines including isoenzyme analysis, human lymphocyte antigen (HLA) typing and STR analysis.
One significant cell-line cross contaminant is the immortal HeLa cell line.
Cell Cultures
Cell cultures are separated into 3 types:-
1. Primary cells - prepared directly from animal or human tissues and can be subcultured only once or twice e.g. primary monkey or baboon kidney
2. Semi-continuous diploid cells - which are derived from human fetal tissue and can be subcultured 20 to 50 times e.g. human diploid fibroblasts such as MRC-5
3. Continuous cells - derived from tumours of human or animal tissue e.g. Vero, Hep2
Cell cultures vary greatly in their susceptibility to different viruses. It is of utmost importance that the most sensitive cell cultures are used for a particular suspected virus. Specimens for cell culture should be transported to the laboratory as soon as possible upon being taken. Swabs should be put in a vial containing virus transport medium. Bodily fluids and tissues should be placed in a sterile container.
Upon receipt, the specimen is inoculated into several different types of cell culture depending on the nature of the specimen and the clinical presentation. The maintenance media should be changed after 1 hour or if that is not practicable, the next morning. The inoculated tubes should be incubated at 35-37oC in a rotating drum. Rotation is optimal for the isolation of respiratory viruses and result in an earlier appearance of the CPE for many viruses. If stationary tubes are used, it is critical that the culture tubes be positioned so that the cell monolayer is bathed in nutrient medium.
The inoculated tubes should be read at least every other day for the presence of cytopathic effect. Certain specimens, such as urine and faeces, may be toxic to cell cultures that may produce a CPE-like effect. If toxic effects are extensive, it may be necessary to passage the inoculated cells. Cell cultures that are contaminated by bacteria should either be put up again or passed through a bacterial filter. Cell cultures should be kept for at least one to two weeks (longer in the case of CMV). Cell cultures should be refed with fresh maintenance medium at regular intervals or if required should the culture medium become too acidic or alkaline. When CPE is seen, it may be advisable to passage infected culture fluid into a fresh culture of the same cell type. For cell-associated viruses such as CMV and VZV, it is necessary to trypsinize and passage intact infected cells. Other viruses such as adenovirus can be subcultured after freezing and thawing infected cells.
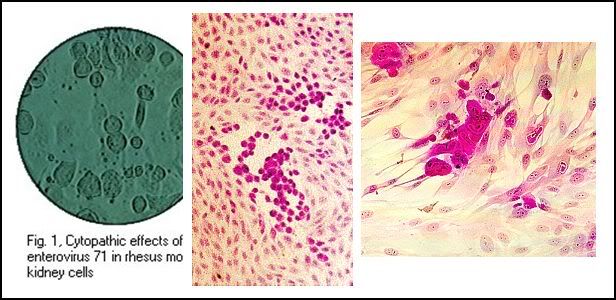
Cytopathic effects of enterovirus 71, HSV, and CMV in cell culture: note the ballooning of cells. (Linda Stannard, University of Cape Town, Virology Laboratory, Yale-New Haven Hospital)
Photo captured from:http://virology-online.com/general/Test1.htm

Cytopathic effects of mumps and measles viruses in cell culture: note the formation of syncytia. (Courtesy of Linda Stannard, University of Cape Town, S.A.)
Photo captured from:http://virology-online.com/general/Test1.htm
Influenza and parainfluenza viruses do not ordinarily induce CPE, however they possess haemagglutinins and thus the ability to absorb guinea pig RBCs as they bud from the cell. This phenomenon is known as haemadsorption.
Commonly employed cell cultures include primary monkey kidney, LLC-MK2 and MDCK cells. The cell cultures are incubated with a suspension of guinea pig RBCs at 4oC or RT for 30 minutes. The unabsorbed RBCs are then removed and the cell sheet observed microscopically for the presence of haemadsorption. Presumptive identification of virus isolates can usually be made on the basis of the type of CPE, haemadsorption, and selective cell culture susceptibility. For final identification, immunofluorescence, neutralization, haemadsorption inhibition, lectron microscopy, or molecular tests are normally carried out.
Advantages of cell culture for virus diagnosis include relative ease, broad spectrum and sensitivity. It is limited by the difficulty in maintaining cell cultures, variability of cell cultures. Contamination by endogenous viral agents such as SV40, mycoplasma and bacteria may occur. Another problem in isolating certain viruses, especially myxo and paramyxo viruses is the presence of inhibitory substances or antibodies in the calf serum used in the cell culture media. Using fetal calf serum reduces this problem but adds to the expense.
Plaque Assay

Photo from:http://www.nanohedron.com/plaque_assay_533.jpg
A plaque is a confluent, monolayer culture of cells. It forms as a result of infection of one of the cells by a single virus particle. Once the cell is infected, the virus replicates and eventually kills the cell. Furthermore, the newly replicated virus particles spread, infect and kill the nearby cells.
In a plaque assay, the culture is first stained with a particular dye. This die stains only viable (live) cells. As a result, the dead cells appear unstained in the culture, against the dyed background. Currently, the plaque assay is applied to detect cells that produce antibodies that destroy erythrocytes by hemolysis.
(Info link: http://www.ehow.com/about_5480231_plaque-assay-method.html)
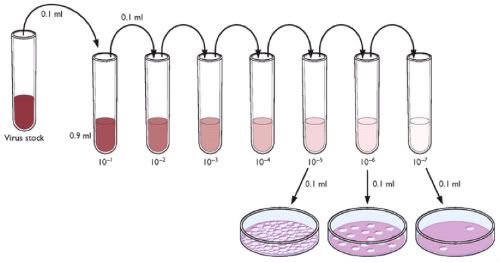
Photo from:http://www.virology.ws/wp-content/uploads/2009/07/plaque-assay.jpg
The plaque assay can be used to purify a clonal population of virus or to determine viral titer as plaque-forming units per ml (pfu/ml) so that known amounts of virus can be used to infect cells during subsequent work. In this assay, cell monolayers are infected with a low ratio of virus, such that sporadic cells become infected. An overlay of agarose keeps the cells stable and limits the spread of virus. When each infected cell produces virus and eventually lyses, only the immediately adjacent cells become infected.
Each group of infected cells is referred to as a plaque. Uninfected cells surround the plaques. After several infection cycles, the infected cells in the center of the plaques begin to lyse and the peripheral infected cells remain surrounded by uninfected cells. This phenomenon causes the light passing through the infected cells to refract differently than the surrounding uninfected cells, and the plaque can be visualized either by the naked eye or by light microscopy.
Each plaque represents a single virus. Therefore, clonal virus populations may be purified by isolating individual plaques. Individual plaques obtained from varying dilutions of a viral stock can be counted to determine the viral titer (pfu/ml) of a given transfection or virus stock. The condition of the cells and their even distribution over the surface of the tissue culture plate is important to the success of a plaque assay. Cells should be healthy, > 95% viable, and in log-phase growth at the time of the assay. Clumpy cells, cells that are not evenly distributed at the correct density (>70%) over the plate, and cells that do not adhere to the tissue culture dishes within 30 min after plating are detrimental to the assay.
(Info link: http://www.bdbiosciences.com/support/resources/baculovirus/plaque_assay.jsp)
Embryonated Eggs

Photo:http://www.studentsguide.in/microbiology/viruses-viroids-virusoids-prions/images/virus-cultivation.jpg
• Still useful in cultivation and detection of some viruses, flu
• Three parts of the egg are of use
1. A: The amniotic cavity:
- Surrounds the embryo & lined by a single layer of epithelial cells.
- The amniotic fluid bathes the external surface of the embryo and comes into contact with the respiratory and alimentary tracts.
2. B: The allantoic cavity:
- Comprises an outgrowth of the hind-gut of the embryo and is lined with endoderm.
• Both are useful for the cultivation of viruses, particularly
- orthomyxoviruses (e.g., influenza) and paramyxoviruses (e.g., mumps).
- Membranes major source of cells in which virus growth occurs, but the embryo may also become infected.
• The allantoic cavity is routinely used because it is technically much simpler to inoculate. However, some viruses (e.g., human influenza isolates) may need to be egg-adapted by growth in the amniotic cavity before they will grow efficiently in the allantoic cavity; the reason for this is not known.
3. C: The chorio-allantoic membrane : The membrane consists of an outer layer of stratified epithelium which constitutes the respiratory surface of the egg, and an inner layer of endoderm (the lining of the allantoic cavity). The membrane may be used as a cell sheet provided it is first dropped away from the shell membrane. Dermatropic viruses (poxviruses and some herpes viruses) will grow on this membrane, and at low concentrations, will give discrete foci of infection which consist of centres of cell proliferation and necrosis (pocks). The membrane may therefore be used to assay these viruses. In addition, different viruses cause pocks of different colour and morphology, and this is of diagnostic value for distinguishing between different poxviruses.
(Info link: www.run.edu.ng/VIRUS%20CULTIVATION%20AND%20IDENTIFICATION.ppt)
Im Chick-Embryo
The animal viruses can be successfully cultivated using chick-embryo technique. In this method fertile hen eggs are selected. Eggs must not be more then 12 days old. To prepared the egg for virus cultivation, the shell surface is first disinfected with iodine and penetrated with a small sterile drill. After inoculation, the drill hole is sealed with gelatin and the egg is then incubated. Viruses may be able to region. For convenience, the mayxoma virus grows well on the chorioallantoic membrane, whereas the mumps virus prefers the allantoic cavity., The infection may produce a local tissue lesion known as pock, whose appearance often is characteristic of the virus.
(Info link: http://www.studentsguide.in/microbiology/viruses-viroids-virusoids-prions/cultivation-of-animal-viruses.html)
---
Detection, Identification and Diagnosis
Under Molecular Biology
Polymerase Chain Reaction
Photo :http://upload.wikimedia.org/wikipedia/commons/8/81/PCR_tubes.png
The polymerase chain reaction (PCR) is a technique to amplify a single or few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence. The method relies on thermal cycling, consisting of cycles of repeated heating and cooling of the reaction for DNA melting and enzymatic replication of the DNA. Primers (short DNA fragments) containing sequences complementary to the target region along with a DNA polymerase (after which the method is named) are key components to enable selective and repeated amplification. As PCR progresses, the DNA generated is itself used as a template for replication, setting in motion a chain reaction in which the DNA template is exponentially amplified. PCR can be extensively modified to perform a wide array of genetic manipulations.
The application of PCR are:
-Selective DNA isolation
-Amplification and quantification of DNA
-PCR in diagnosis of diseases
Enzyme-linked immunosorbent assay (ELISA)
ELISA is a biochemical technique used mainly in immunology to detect the presence of an antibody or an antigen in a sample. The ELISA has been used as a diagnostic tool in medicine and plant pathology, as well as a quality control check in various industries.
In simple terms, in ELISA an unknown amount of antigen is affixed to a surface, and then a specific antibody is washed over the surface so that it can bind to the antigen. This antibody is linked to an enzyme, and in the final step a substance is added that the enzyme can convert to some detectable signal. Thus in the case of fluorescence ELISA, when light of the appropriate wavelength is shone upon the sample, any antigen/antibody complexes will fluoresce so that the amount of antigen in the sample can be inferred through the magnitude of the fluorescence.

Photo :http://www.microvet.arizona.edu/courses/mic419/ToolBox/ELISA2.gif
Performing an ELISA involves at least one antibody with specificity for a particular antigen. The sample with an unknown amount of antigen is immobilized on a solid support (usually a polystyrene microtiter plate) either non-specifically (via adsorption to the surface) or specifically (via capture by another antibody specific to the same antigen, in a "sandwich" ELISA). After the antigen is immobilized the detection antibody is added, forming a complex with the antigen. The detection antibody can be covalently linked to an enzyme, or can itself be detected by a secondary antibody which is linked to an enzyme through bioconjugation.
Applications:
-to evaluate either the presence of antigen or the presence of antibody in a sample
-a useful tool for determining serum antibody concentrations (such as with the HIV test or West Nile Virus)
-detect potential food allergens such as milk, peanuts, walnuts, almonds, and eggs.
-used in toxicology as a rapid presumptive screen for certain classes of drugs.
Under Serological / immunological methods
Western Blot
The western blot (alternatively, protein immunoblot) is an analytical technique used to detect specific proteins in a given sample of tissue homogenate or extract. It uses gel electrophoresis to separate native or denatured proteins by the length of the polypeptide (denaturing conditions) or by the 3-D structure of the protein (native/ non-denaturing conditions). The proteins are then transferred to a membrane (typically nitrocellulose or PVDF), where they are probed (detected) using antibodies specific to the target protein. There are now many reagent companies that specialize in providing antibodies (both monoclonal and polyclonal antibodies) against tens of thousands of different proteins.Commercial antibodies can be expensive, although the unbound antibody can be reused between experiments. This method is used in the fields of molecular biology, biochemistry, immunogenetics and other molecular biology disciplines.
The method originated from the laboratory of George Stark at Stanford. The name western blot was given to the technique by W. Neal Burnette and is a play on the name Southern blot, a technique for DNA detection developed earlier by Edwin Southern. Detection of RNA is termed northern blotting and the detection of post-translational modification of protein is termed eastern blotting.
Haemagglutination
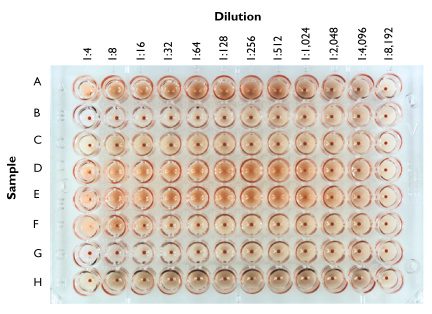
Photo :hhttp://www.virology.ws/wp-content/uploads/2009/05/hemagglutination.jpg
Hemagglutination, or haemagglutination, is a specific form of agglutination that involves red blood cells (RBCs). It has two common uses in the laboratory: blood typing and the quantification of virus dilutions.
Blood Typing
Blood type can be determined by using antibodies that bind to the A or B blood group in a sample of blood.
For example, if antibodies that bind the A blood group are added and agglutination occurs, the blood is either type A or type AB. To determine between type A or type AB, antibodies that bind the B group are added and if agglutination does not occur, the blood is type A.
In blood grouping the patient's serum is tested against RBCs of known blood groups and also the patient's RBCs are tested against known serum types. In this way the patient's blood group is confirmed from both RBCs and serum. A direct Coombs test is also done on the patient's blood sample in case there are any confounding antibodies.
Viral Hemagglutination Assay
Many viruses attach to molecules present on the surface of RBCs. A consequence of this is that at certain concentrations, a viral suspension may bind together (agglutinate) the RBCs, thus preventing them from settling out of suspension. Since agglutination is rarely linked to infectivity, attenuated viruses can therefore be used in assays.
By serially diluting a virus suspension into an assay tray (a series of wells of uniform volume) and adding a standard amount of blood cells, an estimation of the number of virus particles can be made. While less accurate than a plaque assay, it is cheaper and quicker (taking just 30 minutes).
This assay may be modified to include the addition of an antiserum. By using a standard amount of virus, a standard amount of blood cells, and serially diluting the antiserum, one can identify the minimum inhibitory concentration of the antiserum (the greatest dilution which inhibits hemagglutination).
---
Counting of Viruses
Serial dilution
A serial dilution is the stepwise dilution of a substance in solution. Usually the dilution factor at each step is constant, resulting in a geometric progression of the concentration in a logarithmic fashion. A ten-fold serial dilution could be 1 M, 0.1 M, 0.01 M, 0.001 M... Serial dilutions are used to accurately create highly diluted solutions as well as solutions for experiments resulting in concentration curves with a logarithmic scale. A tenfold dilution for each step is called a logarithmic dilution or log-dilution, a 3.16-fold (100.5-fold) dilution is called a half-logarithmic dilution or half-log dilution, and a 1.78-fold (100.25-fold) dilution is called a quarter-logarithmic dilution or quarter-log dilution. Serial dilutions are widely used in experimental sciences, including biochemistry, pharmacology, microbiology, and physics, as well as in homeopathy.
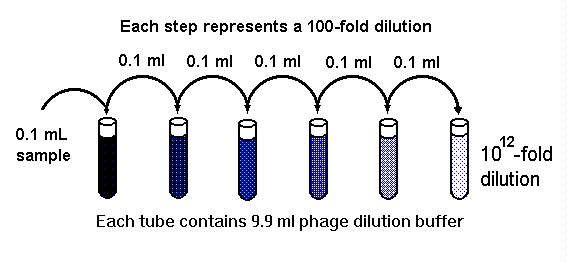
Photo from:http://www.bch.msu.edu/bchug/web/bch472/472lm2c.jpg
Dilution scheme
For a ten-fold logarithmic serial dilution on a 1 ml scale, vials are filled with 1 ml solvent and 0.111 ml solution is serially transferred with thorough mixing after every dilution step. For a half-logarithmic serial dilution, 0.462 ml solution are transferred and for a quarter-log serial dilution 1.285 ml are transferred at each step.
The general formula is: Volume transferred = Volume prefilled / (dilution factor - 1)
Hemocytometer
The hemocytometer or haemocytometer is a device originally designed for the counting of blood cells. It is now also used to count other types of cells as well as other microscopic particles.
The ruled area of the hemocytometer consists of several, large, 1 x 1 mm (1 mm2) squares. These are subdivided in 3 ways; 0.25 x 0.25 mm (0.0625 mm2), 0.25 x 0.20 mm (0.05 mm2) and 0.20 x 0.20 mm (0.04 mm2). The central, 0.20 x 0.20 mm marked, 1 x 1 mm square is further subdivided into 0.05 x 0.05 mm (0.0025 mm2) squares. The raised edges of the hemocytometer hold the coverslip 0.1 mm off the marked grid. This gives each square a defined volume.
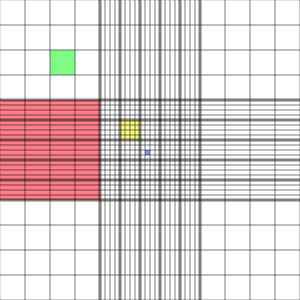
Hemocytometer Grid
red square = 1.0000 mm2, 100.00 nl
green square = 0.0625 mm2, 6.250 nl
yellow square = 0.040 mm2, 4.00 nl
blue square = 0.0025 mm2, 0.25 nl
at a depth of 0.1 mm.
Photo from:http://en.wikipedia.org/wiki/File:Haemocytometer_Grid.png
When a liquid sample containing immobilized cells is placed on the chamber, it is covered with a cover glass, and capillary action completely fills the chamber with the sample. Looking at the chamber through a microscope, the number of cells in the chamber can be determined by counting. Different kinds of cells can be counted separately as long as they are visually distinguishable.
The number of cells in the chamber is used to calculate the concentration or density of the cells in the mixture from which the sample was taken. using the following formula:

Photo captured from:http://en.wikipedia.org/wiki/Hemocytometer
Hemocytometers are often used to count blood corpuscles, organelles within cells, blood cells in cerebrospinal fluid after performing a lumbar puncture, or other cell types in suspension. Using a hemocytometer to count bacteria results in a 'total count' as it is difficult to distinguish between living and dead organisms unless Trypan blue is used to stain the non-viable cells.
|
|

Two more swine flu deaths in Delhi, toll 92
Submitted by Mohit Joshi on Mon, 01/25/2010 - 18:18 swine flu health India New Delhi
New Delhi, Jan 25 :Two more people have died of swine flu in the national capital, raising to 92 the number of fatalities from the viral disease, health authorities said Monday.
Of the two deaths, one is that of a 33-year-old woman and the details of the other casualty is yet to be known.
"A 33-year-old woman died at the Ram Manohar Lohia hospital," Anjana Prakash, deputy nodal officer in charge of swine flu control in Delhi, told IANS.
Besides, the city also reported two fresh cases of swine flu. With these two new cases, the cumulative number of infections in the city has gone up to 9,636. The toll so far is 92 in Delhi.(IANS)
Source: http://www.topnews.in/health/virus-particle-vaccine-could-offer-protection-against-chikungunya-virus-25951
Virus found to replicate four times faster than thought
Submitted by Mohit Joshi on Fri, 01/22/2010 - 11:08 Health News United Kingdom
London, Jan 22 : Live video microscopy shows how vaccinia, a pox virus, spreads four times faster and in a different way than suspected earlier - a discovery that could open the way to the creation of better class of drugs to tackle some viruses.
Vaccinia is a pox virus and is the vaccine that was used to eradicate smallpox. Using live video microscopy, researchers from Imperial College London (ICL) discovered how it spread so fast.
Previously, viruses were thought to spread by entering a cell, replicating there, and then being released to infect new cells, so that the rate of spread of a virus would be limited by how quickly it could replicate in each cell.
Videos of virus-infected cells revealed that the bug spreads by surfing from cell to cell, using a mechanism that allows it to bounce past cells that are already infected and reach uninfected cells as quickly as possible.
Early after vaccinia infects a cell, it expresses two viral proteins on the cell surface, which marks the cell as infected.
When further virus particles reach the infected cell, these proteins cause the host cell to push out snake-like projections called "actin tails," which drive the virus particles away towards other cells that they can infect.
The particles thus bounce from one cell surface to another until they land on an uninfected cell, said an ICL release.
In the study, the researchers prevented the virus from making the proteins needed to make the actin tails in the early stages of infecting a cell and showed that this slowed the spread of the virus dramatically.(IANS)
Source from: http://www.topnews.in/health/virus-found-replicate-four-times-faster-thought-25845
How H1N1 virus infects body
Submitted by Mohit Joshi on Tue, 12/22/2009 - 11:17 Health News United Kingdom
London, Dec 22 : An international team of scientists have made a novel discovery that might help explain how flu virus, including the currently circulating swine-origin H1N1 infects the body.
They have also identified small molecule compounds that act on several of these factors and inhibit viral replication, pointing to new ways to treat flu.
During the study, researchers have identified 295 human cell factors that influenza A strains must harness to infect a cell.
They used RNAi screening technology to selectively turn off more than 19,000 human genes to determine which human factors facilitate viral entry, uncoating, nuclear import, viral replication and other necessary functions of the virus.
"Because influenza mutates so readily, it has become a moving target for therapeutic intervention, making it difficult to treat circulating strains, including the H1N1 swine flu," Nature quoted researcher Sumit Chanda, from Burnham Institute for Medical Research.
"As a result, there is now widespread resistance to two classes of antiviral drugs. However, by targeting more stable human host factors, we may be able to develop therapies that prevent or treat a variety of influenza A strains and are more likely to maintain their effectiveness," Chanda added.
"This study has provided us with crucial knowledge of the cellular pathways and factors the influenza virus exploits to replicate," said co-researcher Megan Shaw, Ph. D., of Mount Sinai.
Each of these represents an ''Achilles heel'' of the virus and vastly increases the number of potential targets for new influenza antiviral drugs, Shaw added.
The team screened human A549 (lung epithelial) cells infected with a modified influenza virus against the genome-wide siRNA library.
The study showed that selectively impairing each of 295 cellular genes reduced viral infection, effectively illuminating the path followed by influenza viruses during the infection of a cell.
Importantly, they found that inhibiting proteins in known drug target classes, such as kinases, vATPases, and tubulin, impairs influenza growth, suggesting that small molecular weight compounds may be developed as host factor-directed antivirals.
These findings were published online in the journal Nature. (ANI)
Source from: http://www.topnews.in/health/how-h1n1-virus-infects-body-25409
Early immune response may help combat hit-and-hide cancer viruses
Submitted by Mohit Joshi on Sat, 01/16/2010 - 12:21 Health News United States
Washington, Jan 16 : Retroviruses such as HIV and HTLV-1 don''t hit-and-run, they hit-and-hide. Now, researchers at Ohio State University have revealed that an early and strong immune response might help curb the infection that is likely to last long.
According to the researchers, cancer viruses slip into host cells and insert their own DNA into the cell''s DNA, and from this refuge they establish an infection that lasts a lifetime.
"Our findings indicate that if the immune system could respond strongly to HTLV-1 and kill infected target cells early, it may inhibit the virus''s ability to establish reservoirs of infected cells and make the infection more manageable later," said principal investigator Michael Lairmore, a professor and chair of veterinary biosciences and a cancer researcher at OSUCCC-James.
"This study tells us that the more we know about the earliest events of infection, the more it will help us develop vaccines and might block those events," Lairemore added.
Lairmore and his colleagues examined HTLV-I infection in rabbits that were treated with the drug cyclosporin A, which is commonly used to suppress the immune system in people following organ transplantation.
They compared animals treated with this drug prior to viral infection with those given the drug one week after infection.
In animals given the immune-suppressing drug first, the virus flourished. The number of virus copies jumped to 200 per 10,000 immune cells (lymphocytes), compared with 40 per 10,000 immune cells in control animals (these were infected with the virus but not given the drug).
After a week or two, the number of virus copies fell, ranging from 113 to 160 for remainder of the 10-week experiment.
In the animals that were given the virus first and then the immune-suppressing drug a week later, on the other hand, the virus languished.
"The first experiment told us that if the immune system is suppressed, the viral load goes up - and we expected that," said Lairmore.
"The second group was the surprise. Their viral load was low from the start, and it stayed that way. We didn''t expect that. We thought the virus would recover and come back up.
"Collectively, our findings indicate that the immune system plays a key role in controlling HTLV-1 spread during early infection, which has important implications for a vaccine against this virus and for therapy for HTLV-1-associated diseases," he added. (ANI)
Source from: http://www.topnews.in/health/early-immune-response-may-help-combat-hit-and-hide-cancer-viruses-25724
|
|

For informations
(For informations that do not fall under the following websites, the links are located right below the informations. )

For informations under "About Virology", "History"
Virology - Wikipedia, the free encyclopedia
Central dogma of molecular biology - Wikipedia, the free encyclopedia
For informations under "Viral replication"
Lytic Cycle - Wikipedia, the free encyclopedia
Lysogenic Cycle - Wikipedia, the free encyclopedia
For informations under "Viral Diseases"
Severe acute respiratory syndrome - Wikipedia, the free encyclopedia
Coronavirus - Wikipedia, the free encyclopedia
HIV - Wikipedia, the free encyclopedia
Enterovirus - Wikipedia, the free encyclopedia
Influenza - Wikipedia, the free encyclopedia
Rubella - Wikipedia, the free encyclopedia
Chickenpox - Wikipedia, the free encyclopedia
Rabies - Wikipedia, the free encyclopedia
Hand, foot and mouth diseases - Wikipedia, the free encyclopedia
Gastroenteritis - Wikipedia, the free encyclopedia
Hepatitis A - Wikipedia, the free encyclopedia
Hepatitis B- Wikipedia, the free encyclopedia
Hepatitis C- Wikipedia, the free encyclopedia
Hepatitis D- Wikipedia, the free encyclopedia
Hepatitis E- Wikipedia, the free encyclopedia
Hepatitis X- Wikipedia, the free encyclopedia
For informations under "Virus Structure"
Virus - Wikipedia, the free encyclopedia
For informations under "Culturing Viruses"
ELISA - Wikipedia, the free encyclopedia
Western blot - Wikipedia, the free encyclopedia
Polymerase chain reaction - Wikipedia, the free encyclopedia
Hemagglutination - Wikipedia, the free encyclopedia
Serial dilution - Wikipedia, the free encyclopedia
Hemocytometer- Wikipedia, the free encyclopedia
Cell culture- Wikipedia, the free encyclopedia

Wong's Virology
For News

Top news | Health news and Updates
For videos
Youtube - Broadcast Yourself
Recommended Websites

All the Virology on the WWW

virology blog — About viruses and viral disease
|
|
This HTML form was created by Freedback.
Browser: Mozilla Firefox
Screen resolution: 1280 x 800 pixels
Established since: DD Month YYYY
Host: Blogger
Layout: Indie Devotee
Done by: Victoria
|